MMCIF (.cif)
Background & Context

-
- MIME types: chemical/x-cif, chemical/x-mmcif
- 3D molecular model file.
- Used in cheminformatics applications and on the web for storing and exchanging molecule models.
- Commonly used as an alternative to the PDB format.
- mmCIF is an acronym derived from Macromolecular Crystallographic Information File.
- Derived from the CIF file format.
- Plain text format.
- Stores structure information for large biological molecules such as proteins and nucleic acids.
- Developed between 1990 and 2005 by the International Union of Crystallography.
Import & Export
- Import["file.cif","MMCIF"] reads an mmCIF file and returns a symbolic representation of a biomolecule.
- The Wolfram Language provides a variety of 3D rendering styles for macromolecules.
- Export["file.cif", biomol] creates an MMCIF file from a biomolecule.
- Import["file.cif","MMCIF"] returns a BioMolecule object.
- Import["file.cif",{"MMCIF",elem}] imports the specified element from an MMCIF file.
- Import["file.cif",{"MMCIF",elem,suba,subb,…}] imports a subelement.
- Import["file.cif",{"MMCIF"{elem1,elem2,…}}] imports multiple elements.
- Export["file.cif",biomol] exports the BioMolecule biomol.
- See the following reference pages for full general information:
-
Import, Export import from or export to a file CloudImport, CloudExport import from or export to a cloud object ImportString, ExportString import from or export to a string ImportByteArray, ExportByteArray import from or export to a byte array
Import Elements



- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - Data elements:
-
"BioMolecule" a symbolic representation of the macromolecular model "Molecule" a symbolic representation of the molecular model - Import and Export use the "BioMolecule" element by default for the mmCIF format.
- A BioMolecule object contains information about the chains and residues, as well as atom types and coordinates. A Molecule object will assign bonds between atoms and discards metainformation such as residue and chain labels.
- Graphics element:
-
"Graphics3D" mmCIF file rendered as a Graphics3D object - Data representation elements:
-
"Residues" residue sequences as an array of three-letter abbreviations "Sequence" residue sequences given as a list of strings "ResidueAtoms" list of residue atoms "ResidueChainLabels" list of chain labels "ResidueRoles" functional roles of residue atoms "ResidueCoordinates" 3D coordinates of residue atoms in angstroms "Resolution" spatial resolution of the model coordinates in angstroms "AdditionalAtoms" atoms that are not constituents of a chain "AdditionalCoordinates" 3D coordinates of additional atoms "AdditionalResidues" additional residue sequences as an array of three-letter abbreviations "SecondaryStructure" rules describing the large-scale structure of a chain "VertexCoordinates" atomic coordinates, given in angstroms "VertexTypes" all atoms or groups constituting the molecule, typically given as a list of chemical element abbreviations - The Wolfram Language uses the standard IUB/IUPAC abbreviations for amino acid residues.
- When importing an mmCIF file that describes multiple 3D models of the same molecule, the following Import elements can be used to read the geometries of all models:
-
"ResidueCoordinatesList" residue coordinates for each model "AdditionalCoordinatesList" 3D coordinates of additional atoms for each model "VertexCoordinatesList" atomic coordinates for each model, in angstroms - Meta-information elements:
-
"Authors" author information as referenced in the file "DepositionDate" when the file was added to the database "PDBClassification" PDB classification from the file header "PDBID PDB structure identification string "References" bibliographic reference, given as a list of rules "Title" document title
Options

- The "BioMolecule" import element takes the following options:
-
"DetectSecondaryStructure" Automatic whether to scan the list of residues to detect helices and sheets - The "Graphics3D" import element takes the same options as BioMoleculePlot3D.
- Selecting a rendering style:
-
PlotTheme "Ribbons" specifies the visualization method - Supported plot themes include:
-

"Ribbons" display polymer chains as ribbons 
"Backbone" display polymer chains as ribbons 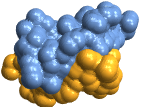
"SolventAccessibleSurface" solvent accessible surface 
"GaussianSurface" Gaussian surface 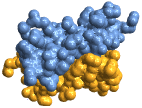
"VanDerWaalsSurface" van der Waals surface 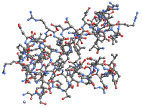
"BallAndStick" display atoms and bonds using Sphere and Cylinder primitives 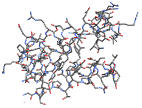
"Tubes" display bonds as tubes with no atoms 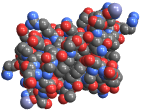
"Spacefilling" atoms are depicted with spheres with radius matching the van der Waals radius
Examples
Basic Examples (4)
Show the names of all available Import elements:
Read reference information from this mmCIF file:
Import the amino acid sequences as strings:
Show the Import elements available in a sample file:
Read all data from an mmCIF file and export it back to the same format:
Export a Molecule object as an mmCIF file: