MoleculeValue[molecule,property]
gives the value of the specified property for the given molecule.
MoleculeValue[{molecule1,molecule2,…},property]
gives the list of values for the specified property for each of the moleculei.
MoleculeValue[molecule,{property1,property2,…}]
gives the list of values of the propertyi for the specified molecule.
MoleculeValue[molecule,{property,item}]
gives the value of the specified property for item in molecule.
MoleculeValue[{molecule1,molecule2,…},{property1,property2,…}]
gives the list of values of the propertyi for each of the moleculei.




MoleculeValue
MoleculeValue[molecule,property]
gives the value of the specified property for the given molecule.
MoleculeValue[{molecule1,molecule2,…},property]
gives the list of values for the specified property for each of the moleculei.
MoleculeValue[molecule,{property1,property2,…}]
gives the list of values of the propertyi for the specified molecule.
MoleculeValue[molecule,{property,item}]
gives the value of the specified property for item in molecule.
MoleculeValue[{molecule1,molecule2,…},{property1,property2,…}]
gives the list of values of the propertyi for each of the moleculei.
Details and Options

















- molecule[property] is equivalent to MoleculeValue[molecule,property] when molecule is a valid molecule.
- MoleculeValue["Properties"] returns all available properties.
- When requesting a property value that depends on atomic coordinates, those coordinates will be computed automatically if not already present in the molecule. If no coordinates can be provided, a Missing value will be returned.
- For properties referring to parts of a molecule, item can have the following forms:
-
ind one atom index (1,2,…) {ind1,ind2,…} a list of atom indices {{ind1,ind2},…} a list of lists of atom indices bnd one bond (Bond[{ind1,ind2},type]) {bnd1,bnd2,…} a list of bonds All all atoms or bonds - General molecule properties include:
-
"AtomDiagramCoordinates" structure diagram coordinates "BridgeheadAtoms" atoms common to rings sharing at least two bonds "ElementCounts" association <|el->n,…|> of "Element" entities and the corresponding number of atoms "ElementMassFraction" association <|el->n,…|> of "Element" entities and their corresponding mass fraction as a percentage "ElementTally" list {{el,n},…} of "Element" entities and the corresponding number of atoms "ExplicitAtomCount" number of atoms in the molecule, excluding implicit hydrogens "ExplicitAtomList" list of atoms in the molecule, excluding implicit hydrogens "ExplicitBondCount" number of bonds in the molecule, excluding implicit hydrogens "ExplicitBondList" list of bonds in the molecule, excluding implicit hydrogens "FullAtomCount" number of atoms in the molecule "FullAtomList" list of atoms in the molecule "FullBondCount" number of bonds in the molecule "FullBondList" list of bonds in the molecule "MetaInformation" any metainformation contained in the molecule expression "MMFFEnergy" energy computed using the MMFF94 force field "MMFFsEnergy" energy computed using the MMFF94s force field "MolarMass" molar mass "MolecularMass" molecular mass, using the average atomic mass for atoms with no specified mass number "MonoIsotopicMolecularMass" molecular mass using the most abundant isotope for all atoms "PossibleStereocenters" list of atom indices for chiral atoms "RelativeMolecularMass" ratio of the molecular mass to the unified atomic mass unit "ResonanceStructureList" list of molecule resonance structures "SpiroAtoms" atoms shared between rings that share exactly one atom "StereochemistryElements" a list of associations detailing any defined local stereochemistry as defined by StereochemistryElements "TautomerList" list of molecule tautomers "TotalCharge" sum of all formal charges in the molecule "UFFEnergy" energy computed using the UFF force field - Properties corresponding to a molecule identifier include:
-
"InChI" international chemical identifier "InChIKey" international chemical identifier key "MolecularFormulaString" molecular formula string "MolecularFormula" molecular formula in display form "SMILES" SMILES string {"SMILES",{id1,id2,…}} SMILES string containing the specified atoms and rooted at the atom with index id1 "CanonicalSMILES" SMILES string with atoms in canonical order, with hydrogens made implicit "IsomericSMILES" SMILES string with atoms in canonical order, with implicit hydrogens, with isotope and stereo information included "AnonymousGraphSMILES" SMILES string with all atoms set to "*" and all single bonds "ElementGraphSMILES" SMILES string with all bonds set to single "MurckoScaffoldSMILES" SMILES string after removing substituents "ExtendedMurckoScaffoldSMILES" SMILES string replacing substituents with "*" "PubChemSynonyms" list of molecule alternate names retrieved from "PubChem" - The "SMILES" property will include all explicit hydrogen atoms present in the molecule, and does not canonicalize the atom order.
- Properties returning a list of ExternalIdentifier objects include:
-
"CASRegistryNumber" CAS registry number "ChEMBLID" ChEMBL identification number "ChemSpiderID" ChemSpider identification number "PubChemCompoundID" PubChem compound ID "PubChemSubstanceID" PubChem substance ID "WikidataID" Wikidata ID - Internet connectivity is required to retrieve external identifiers for a molecule.
- Properties referring to an atom within a molecule may be specified via {property,id} or {property,{id1,id2,…}}, where id is an atom index.
- Per-atom properties include:
-
"AromaticAtomQ" gives True if an atom is aromatic "AtomChirality" absolute atomic chirality; returns "R", "S", "r", "s", or "Unspecified" for chiral atoms and None otherwise "AtomicMass" atomic mass "AtomicNumber" atomic number "AtomicSymbol" IUPAC atomic symbol "AtomIndex" atom index "AtomSMILES" atomic "SMILES" symbol "AtomSMARTS" atomic "SMARTS" symbol "CIPRank" atom rank using the Cahn–Ingold–Prelog priority rules "CoordinationNumber" number of bonded atoms "DefaultValence" default valence "Element" the corresponding "Element" entity "FormalCharge" formal charge "GasteigerPartialCharge" atomic charge using the Gasteiger charge model "GeometricStericEffectIndex" geometric steric index; requires atomic coordinates "HeavyAtomCoordinationNumber" number of bonds to heavy atoms "HydrogenCount" number of hydrogens bonded to the atom "ImplicitHydrogenCount" number of implict hydrogens bonded to the atom "Isotope" the corresponding "Isotope" entity "MassNumber" mass number; returns None if not specified "MMFFPartialCharge" atomic charge computed using the MMFF force field "MostAbundantMassNumber" mass number for the most abundant isotope "OrbitalHybridization" computed orbital hybridization "OuterShellElectronCount" number of electrons in the outer shell "PiElectronCount" number of 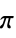 electrons
electrons"RingAtomQ" gives True if an atom is part of a ring "TopologicalStericEffectIndex" topological steric effect index "UnpairedElectronCount" number of unpaired electrons "UnsaturatedAtomQ" gives True if an atom is unsaturated "Valence" number of valence electrons - Properties referring to a bond within a molecule may be specified via {property,bnd} or {property,{bnd1,bnd2,…}}, where bnd is a Bond object or a list of two bonded atom indices.
- Per-bond properties include:
-
"BondIndex" position of a bond in the bond list for a molecule "BondLength" Euclidean distance between the given atoms "BondOrder" numerical bond order "BondStereo" absolute bond stereo as determined using the Cahn–Ingold–Prelog priority rules "BondType" bond type, e.g. "Single", "Double", etc. "ConjugatedBondQ" gives True if the bond is part of a conjugated system "RingBondQ" gives True if the bond is part of a ring - For an atom or bond property, property is equivalent to {property,All}.
- With the default option setting IncludeHydrogensAll, property values for all atoms will be returned by MoleculeValue. Use the option value "ExplicitOnly" to exclude implicit hydrogens.
- Molecular graph properties include:
-
"AdjacencyMatrix" adjacency matrix "BondWeightedAdjacencyMatrix" bond-weighted adjacency matrix "BurdenMatrix" classical Burden matrix "GraphDistanceMatrix" graph distance matrix "SmallestSetOfSmallestRings" atom indices for the smallest set of smallest rings - Properties that depend on molecule geometry include:
-
"AtomCoordinates" list of Cartesian atomic coordinates {"BondAngle",{id1,id1,id3}} the planar angle formed by the three atoms with specified indices "CenterOfMass" center of mass {"CenterOfMass",{id1,id2,…}} center of mass including only the listed atoms "CoulombMatrix" Coulomb matrix, with elements 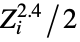 and
and ![Z_i Z_j/TemplateBox[{{{R, _, i}, -, {R, _, j}}}, Abs] Z_i Z_j/TemplateBox[{{{R, _, i}, -, {R, _, j}}}, Abs]](Files/MoleculeValue.en/3.png) where
where 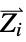 and
and 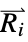 are the nuclear charges and positions
are the nuclear charges and positions"CoulombMatrixEigenvalues" eigenvalues of the Coulomb matrix "DistanceMatrix" matrix of Euclidean distances between atom centers "InertiaTensor" inertia tensor {"InteratomicDistance",{id1,id2}} Euclidean distance between atoms with the given indices {"OutOfPlaneAngle",{id1,id2,id3,id4}} Wilson angle between the plane containing atoms id1, id2 and id3 and the bond containing atoms id2 and id4 "PrincipalAxes" principal axes, eigenvectors of the molecule's inertia tensor "PrincipalMoments" principal moments of inertia {"TorsionAngle",{id1,id2,id3,id4}} torsion angle between the plane containing atoms id1, id2 and id3 and the plane containing atoms id2, id3 and id4 - Geometric properties "BondAngle", "InteratomicDistance", "OutOfPlaneAngle" and "TorsionAngle" may also take a list of lists of atom indices, in which case a list of values is returned.
- Properties pertaining to molecular symmetry include:
-
"PointGroupDisplay" Schoenflies notation for the molecular point group in display form "PointGroupString" molecular point group as a string "SymmetryElements" a list of the symmetry elements including rotation axes, planes of symmetry and inversion centers - Available topological descriptors include:
-
"AliphaticCarbocycleCount" aliphatic carbocycle count "AliphaticHeterocycleCount" aliphatic heterocycle count "AliphaticRingCount" aliphatic ring count "AmideBondCount" amide bond count "AromaticCarbocycleCount" aromatic carbocycle count "AromaticHeterocycleCount" aromatic heterocycle count "AromaticMoleculeQ" returns True if the molecule contains aromatic bonds "AromaticRingCount" aromatic ring count "Autocorrelation2D" 2D autocorrelation "BoettcherComplexity" Böttcher's molecular complexity "BridgeheadAtomCount" number of bridgehead atoms, atoms common to rings sharing at least two bonds "Chi0n"-"Chi4n" Kier and Hall connectivity indices "Chi0v"-"Chi4v" Kier and Hall valence connectivity indices "CrippenClogP" Crippen calculated log P "CrippenMR" Crippen molar refactivity "DegreeOfUnsaturation" degree of unsaturation "FractionCarbonSP3" fraction of carbon atoms with sp3 hybridization "HBondAcceptorCount" hydrogen-bond acceptor count "HBondDonorCount" hydrogen-bond donor count "HeteroatomCount" heteroatom count "HeterocycleCount" heterocycle count "Kappa1"-"Kappa3" κ shape indices "KierHallAlphaShape" Kier and Hall modified shape index "LabuteApproximateSurfaceArea" Labute approximate surface area "LipinskiHBondAcceptorCount" Lipinski H-bond acceptor count "LipinskiHBondDonorCount" Lipinski H-bond donor count "MolecularQuantumNumbers" molecular quantum numbers "PEOEVSA" partial equalization of orbital electronegativity van der Waals surface area "QuantitativeEstimateOfDrugLikeness" quantitative estimation of drug‐likeness "RingCount" ring count "RotatableBondCount" rotatable bond count "SaturatedCarbocycleCount" saturated carbocycle count "SaturatedHeterocycleCount" saturated heterocycle count "SaturatedRingCount" saturated ring count "SlogPVSA" Crippen log P van der Waals surface area "SMRVSA" Crippen molar refractivity van der Waals surface area "SpiroAtomCount" spiro atom count "StereocenterCount" the number of possible stereocenters "SyntheticAccessibilityScore" synthetic accessibility score "TopologicalPolarSurfaceArea" topological polar surface area "UnspecifiedStereocenterCount" unspecified stereocenter count - Geometric molecular descriptors, which depend on the atomic coordinates, include:
-
"Asphericity" asphericity, defined by 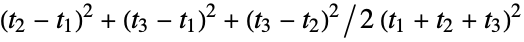 where
where 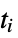 are the moments of the gyration tensor
are the moments of the gyration tensor"Autocorrelation3D" 3D autocorrelation "Eccentricity" eccentricity, defined by 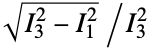 where
where 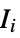 are the moments of the inertia tensor
are the moments of the inertia tensor"GETAWAY" geometry, topology and atom-weights assembly descriptors "GETAWAYAssociation" Association version of the "GETAWAY" property "InertialShapeFactor" inertial shape factor, defined as 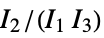
"MORSE" 3D‐molecule representation of structures based on electron diffraction "MORSEAssociation" Association version of the "MORSE" property "NormalizedPrincipalMomentRatios" first normalized principal moments ratio, { 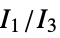 ,
,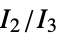 }
}"PlaneOfBestFitDistance" average distance from heavy atoms to the plane of best fit "RadiusOfGyration" radius of gyration, 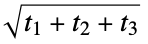
"RDF" radial distribution function descriptors "SpherosityIndex" spherosity index, defined as 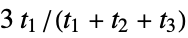
"WHIM" weighted holistic invariant molecular descriptors "WHIMAssociation" Association version of the "WHIM" property - The following modifiers can be used in MoleculeValue[molecule,property,"modifier"]:
-
"PropertyAssociation" an association of properties and property values "Dataset" a dataset where the rows are associations of properties and property values "MoleculePropertyAssociation" an association in which the specified molecules are keys, and values are a nested association of properties and property values - When applicable, MoleculeValue[ MoleculeProperty[property],"MetaInformation"] will provide a dataset of available information, such as a citation.
Examples
open all close allBasic Examples (4)
Scope (14)
3D Descriptors (2)
Atom Properties (3)
Bond Properties (2)
Geometric Properties (2)
Geometric properties depend on a molecule's 3D coordinates. If none are provided, automatic coordinates are generated. Create a molecule from a "Chemical" entity and use the coordinates from the Wolfram Knowledgebase:
Find the distance between two atoms and highlight them in a 3D plot:
Find the out-of-plane angle for four atoms:
When no atom indices are specified, geometric properties will return an association whose keys are the atom indices:
Graph Properties (1)
Topological Descriptors (2)
List the topological descriptors:
Find the fraction of carbon atoms with sp3 hybridization for a list of molecules:
The "BridgeheadAtomCount" returns the number of atoms common to rings that share at least two bonds:
Use the "BridgeheadAtoms" property to visualize the bridgehead atoms:
Note that this definition includes only bridged compounds and excludes simple fused systems:
For a count that includes fused-ring atoms, use an Atom pattern:
Identifiers (2)
The "SMILES" property will contain all hydrogen atoms and makes no attempt to canonicalize atom order. "CanonicalSMILES" uses canonical ordering and implicit hydrogen atoms, but removes isotope and stereo information. "IsomericSMILES" is a canonical version with stereo and isotopes. Compare the three SMILES properties with a Dataset:
Find the InChIKey for a molecule:
Search for this molecule using WikidataData:
Options (2)
IncludeHydrogens (2)
When querying a per-atom property, values for all atoms are included:
Use IncludeHydrogensNone to exclude the hydrogens, which make up the majority of atoms:
Hydrogen atoms can often be omitted when their presence can be inferred by normal valence rules. Use IncludeHydrogens"ExplicitOnly" to include only the atoms listed explicitly:
Possible Issues (1)
Because Molecule will try to fill valences with hydrogen atoms, hydrogen atoms will be included in the results even if they were not in the original molecule expression:
To exclude implicit hydrogen atoms, use:
Alternatively, to stop hydrogen atoms from appearing, use the Molecule option ValenceFillingNone:
See Also
Molecule MoleculeProperty IncludeHydrogens BondList AtomList
Function Repository: MoleculeValuePlot MoleculeValuePlot3D MolecularComplexity MACCSKeys SmilesString
Related Guides
Related Links
History
Introduced in 2019 (12.0) | Updated in 2020 (12.1) ▪ 2020 (12.2) ▪ 2021 (12.3) ▪ 2022 (13.1)
Text
Wolfram Research (2019), MoleculeValue, Wolfram Language function, https://reference.wolfram.com/language/ref/MoleculeValue.html (updated 2022).
CMS
Wolfram Language. 2019. "MoleculeValue." Wolfram Language & System Documentation Center. Wolfram Research. Last Modified 2022. https://reference.wolfram.com/language/ref/MoleculeValue.html.
APA
Wolfram Language. (2019). MoleculeValue. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/MoleculeValue.html
BibTeX
@misc{reference.wolfram_2025_moleculevalue, author="Wolfram Research", title="{MoleculeValue}", year="2022", howpublished="\url{https://reference.wolfram.com/language/ref/MoleculeValue.html}", note=[Accessed: 24-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_moleculevalue, organization={Wolfram Research}, title={MoleculeValue}, year={2022}, url={https://reference.wolfram.com/language/ref/MoleculeValue.html}, note=[Accessed: 24-February-2026]}