CML (.cml)
Background & Context
-
- MIME type: chemical/x-cml
- Chemical Markup Language, for interchange of chemical information.
- Represents an arbitrary number of chemical compounds.
- XML format.
- Stores atomic coordinates, chemical bond information and metadata.
Import & Export

- Import["file.cml"] imports a list of molecules from a CML file.
- Export["file.cml",expr] exports a molecule or list of molecules to a CML file.
- Import["file.cml",elem] imports the specified element from a CML file.
- The import format can be specified with Import["file","CML"] or Import["file",{"CML",elem,…}].
- Export["file.cml",mol] creates a CML file from a molecule.
- Export["file.cml",{mol1,mol2,…}] creates a CML file from a list of molecules.
- See the following reference pages for full general information:
-
Import, Export import from or export to a file CloudImport, CloudExport import from or export to a cloud object ImportString, ExportString import from or export to a string ImportByteArray, ExportByteArray import from or export to a byte array
Import Elements

- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - Data elements:
-
"Molecule" a symbolic representation of the molecule model "Molecule", n a symbolic representation of the n 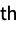 molecule
molecule"Metadata" an Association containing the metadata from the file "Metadata", n the metadata for the n 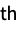 molecule
molecule
Options
- General Import options:
-
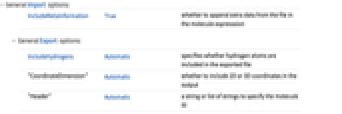
IncludeMetaInformation True whether to append extra data from the file in the molecule expresssion - General Export options:
-
IncludeHydrogens Automatic specifies whether hydrogen atoms are included in the exported file "CoordinateDimension" Automatic whether to include 2D or 3D coordinates in the output "Header" Automatic a string or list of strings to specify the molecule ID
Examples
open all close allBasic Examples (2)
Show the Import elements available in a CML file:
Import all molecules from the file:
Import the second and fifth molecules from the file:
Export Options (2)
IncludeHydrogens (1)
With the default setting of IncludeHydrogensAutomatic, hydrogen atoms are included in the output file if they are explicitly present in the molecule:
Specify that explicit hydrogens should be added before export:
"CoordinateDimension" (1)
The option "CoordinateDimension" can take values of Automatic, 2 or 3.
Export a molecule to CML with two-dimensional coordinates:
Export a molecule to CML with three-dimensional coordinates:
History
Introduced in 2020 (12.1)