SMILES (.smi)
SMILES (.smi)
Background & Context

-
- MIME type: chemical/x-daylight-smiles
- SMILES chemical format.
- Commonly used to describe the structure of chemical molecules.
- SMILES is an acronym for Simplified Molecular Input Line Entry Specification.
- Used in cheminformatics applications and in chemistry databases to represent chemical formulas.
- ASCII format.
- Uses a linear notation to represent the connectivity graph of a molecule.
- Can store data for multiple molecules.
- Additional properties can be stored on the same line as the SMILES string.
- Developed in the 1980s by Arthur Weininger and David Weininger.
Import & Export
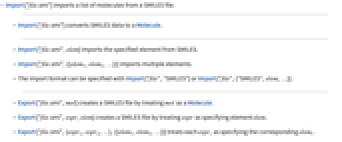
- Import["file.smi"] imports a list of molecules from a SMILES file.
- Import["file.smi"] converts SMILES data to a Molecule.
- Import["file.smi",elem] imports the specified element from SMILES.
- Import["file.smi",{{elem1,elem2,…}}] imports multiple elements.
- The import format can be specified with Import["file","SMILES"] or Import["file",{"SMILES",elem,…}].
- Export["file.smi",mol] creates a SMILES file by treating mol as a Molecule.
- Export["file.smi",expr,elem] creates a SMILES file by treating expr as specifying element elem.
- Export["file.smi",{expr1,expr2,…},{{elem1,elem2,…}}] treats each expri as specifying the corresponding elemi.
- Export["file.smi",expr,opt1->val1,…] exports expr with the specified option elements taken to have the specified values.
- Export["file.smi",{elem1->expr1,elem2->expr2,…},"Rules"] uses rules to specify the elements to be exported.
- See the following reference pages for full general information:
-
Import, Export import from or export to a file CloudImport, CloudExport import from or export to a cloud object ImportString, ExportString import from or export to a string ImportByteArray, ExportByteArray import from or export to a byte array
Import Elements

- General Import elements:
-
"Elements" list of elements and options available in this file "Summary" summary of the file "Rules" list of rules for all available elements - Data elements:
-
"Molecule" a symbolic representation of the molecule model "Molecule", n a symbolic representation of the n 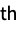 molecule
molecule"Metadata" an Association containing the metadata from the file "Metadata", n the metadata for the n 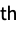 molecule
molecule - Import by default uses the "Molecule" element when converting from SMILES.
- SMILES strings are interpreted by Molecule.
- Data representation elements:
-
"EdgeRules" connectivity data, given as a list of rules "EdgeTypes" bond types, given as a list of strings "FormalCharges" charges of the atoms given by "VertexTypes" "VertexTypes" all atoms or groups constituting the molecule, typically given as a list of chemical element abbreviations
Examples
Basic Examples (6)
Import the SMILES string for caffeine:
Find the corresponding chemical entity:
Import a list of molecules from a SMILES file string:
Import a list of molecules along with meta information:
The metadata is stored in the molecule expressions:
Export a list of molecules as SMILES strings:
Include a data column in the output:
Use associations for the metadata to generate a header line:
Import from a SMILES string, keeping hydrogen atoms implicit:
Related Guides
History
Introduced in 2008 (7.0) | Updated in 2019 (12.0)