BioSequence[type,"seq"]
represents the biomolecular sequence of the given type corresponding to a string "seq".
BioSequence["seq"]
infers the type (DNA, protein, etc.) from the sequence.
BioSequence[ent]
gives the biomolecular sequence associated with the gene or protein entity ent.
BioSequence[type,{chem1,chem2,…}]
gives the biomolecular sequence with type corresponding to the given list of chemicals.
BioSequence[type,"seq",{bond1,bond2,…}]
represents a biomolecular sequence with the given list of bonds.
BioSequence["HybridStrand",{bioseq1,bioseq2,…},{bond1,bond2,…}]
represents a sequence composed of multiple motif sequences with shared primary linkage.
BioSequence[{bioseq1,bioseq2,…},{bond1,bond2,…}]
represents a number of sequences linked only by additional bonds.


BioSequence
BioSequence[type,"seq"]
represents the biomolecular sequence of the given type corresponding to a string "seq".
BioSequence["seq"]
infers the type (DNA, protein, etc.) from the sequence.
BioSequence[ent]
gives the biomolecular sequence associated with the gene or protein entity ent.
BioSequence[type,{chem1,chem2,…}]
gives the biomolecular sequence with type corresponding to the given list of chemicals.
BioSequence[type,"seq",{bond1,bond2,…}]
represents a biomolecular sequence with the given list of bonds.
BioSequence["HybridStrand",{bioseq1,bioseq2,…},{bond1,bond2,…}]
represents a sequence composed of multiple motif sequences with shared primary linkage.
BioSequence[{bioseq1,bioseq2,…},{bond1,bond2,…}]
represents a number of sequences linked only by additional bonds.
Details and Options











- BioSequence[…] evaluates, if possible, to the following forms:
-
BioSequence[type,"seq",bonds] motifs (single strands of a single type) BioSequence["HybridStrand",{bioseq1,bioseq2,…},bonds] hybrid strands (single strands of multiple types) BioSequence[{bioseq1,bioseq2,…},bonds] sequence collections (many strands with additional bonds) - BioSequence employs the following letters to represent molecules for each type:
-
"DNA" A, C, G, T "CircularDNA" A, C, G, T "RNA" A, C, G, U "CircularRNA" A, C, G, U "Peptide" A, C, D, E, F, G, H, I, K, L, M, N, O, P, Q, R, S, T, U, V, W, Y "CircularPeptide" A, C, D, E, F, G, H, I, K, L, M, N, O, P, Q, R, S, T, U, V, W, Y - The content of this table is available through the "Alphabet" property of "BioSequenceType" entities; for example through Entity["BioSequenceType","DNA"]["Alphabet"].
- Here is the corresponding nucleotide for each DNA (RNA) letter:
-
A adenine C cytosine G guanine T (U) thymine (uracil) - Similarly, here is the corresponding amino acid for each peptide letter:
-
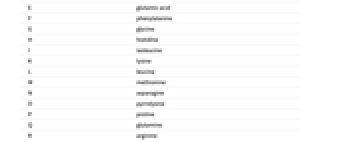
A alanine C cysteine D aspartic acid E glutamic acid F phenylalanine G glycine H histidine I isoleucine K lysine L leucine M methionine N asparagine O pyrrolysine P proline Q glutamine R arginine S serine T threonine U selenocysteine V valine W tryptophan Y tyrosine - The content of the previous tables is available through the "AlphabetRules" property of "BioSequenceType" entities, for example through Entity["BioSequenceType","DNA"]["AlphabetRules"].
- The "Peptide" and "CircularPeptide" types also allow a period or asterisk (. or *) to represent where a stop in biomolecular translation occurs.
- Additionally, the type can be None to represent generic sequences with no given chemical meaning.
- BioSequence also allows degenerate letters that represent a number of potential chemicals.
- Allowed degenerate letters for DNA and RNA include:
-
B C, G or T/U (not A) D A, G or T/U (not C) H A, C or T/U (not G) K G or T/U (keto) M A or C (amino) N A, C, G or T/U (any letter) R A or G (purine) S C or G (strong) V A, C or G (not T) W A or T/U (weak) Y C or T/U (pyrimidine) - Allowed degenerate letters for peptides include:
-
B D or N J I or L X A, C, D, E, F, G, H, I, K, L, M, N, O, P, Q, R, S, T, U, V, W, Y Z E or Q - The content of the previous tables is available through the "DegenerateLetterRules" property of "BioSequenceType" entities, e.g. Entity["BioSequenceType","DNA"]["DegenerateLetterRules"].
- The following letter is used as the arbitrary letter when a type and length are provided:
-
"DNA" or "CircularDNA" N "RNA" or "CircularRNA" N "Peptide" or "CircularPeptide" X - BioSequence accepts standard abbreviations in place of sequence letters.
- Possible abbreviations for DNA bases include:
-
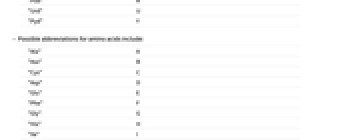
"dAdo" A "dCyd" C "dGuo" G "dNuc" N "dPuo" R "dThd" T "dPyd" Y - Possible abbreviations for RNA bases include:
-
"Ado" A "Cyd" C "Guo" G "Nuc" N "Puo" R "Urd" U "Pyd" Y - Possible abbreviations for amino acids include:
-
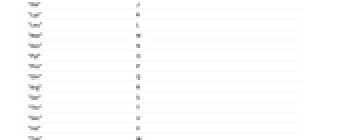
"Ala" A "Asx" B "Cys" C "Asp" D "Glu" E "Phe" F "Gly" G "His" H "Ile" I "Xle" J "Lys" K "Leu" L "Met" M "Asn" N "Pyl" O "Pro" P "Gln" Q "Arg" R "Ser" S "Thr" T "Sec" U "Val" V "Trp" W "Xaa" X "Tyr" Y "Glx" Z - In addition to the connections implied by the sequence, BioSequence letters can be connected through additional Bond entries.
- Bonds specified in the form Bond[{i,j},type] connect the chemicals corresponding to the string positions i and j through a bond of type type. For example, the hydrogen bonds connecting the "A" and the "T" in the DNA sequence "ACCT" could be represented as BioSequence["DNA","ACCT",{Bond[{1,4},"MultiHydrogen"]}].
- A single bond at the sequence level can represent multiple bonds at the molecular level. In the previous example, the Bond between the "A" and the "T" represents two hydrogen bonds at the molecular level.
- In a hybrid strand, bonds of the form Bond[{{i1,i2},{j1,j2}},type] connect the motif strands with indices i1 and j1 at positions i2 and j2, respectively, through a bond of the specified type. For example, the hydrogen bonds connecting the "A" and the "U" in the DNA/RNA hybrid sequence {"ACC","CCU"} could be represented as BioSequence["HybridStrand",{"ACC","CCU"},{Bond[{{1,2},{2,3}},"MultiHydrogen"]}].
- In a sequence collection, bonds of the form Bond[{{i1,i2,i3},{j1,j2, j3}},type] connect the motif strands with indices {i1,i2} and {j1,j2} at positions i3 and j3, respectively, through a bond of type type.
- If motif strands are being connected at the sequence collection level, either {i1,1,i3} or {i1,i3} may be used. For example, given two DNA sequences "CAC" and "CTC", the hydrogen bonds connecting the "A" of the first sequence and the "T" of the second sequence can be represented as either BioSequence[{"CAC","CTC"},Bond[{{1,1,2},{2,1,2}},"MultiHydrogen"]] or BioSequence[{"CAC","CTC"},Bond[{{1,2},{2,2}},"MultiHydrogen"]] .
- For a hybrid strand in a sequence collection, all indexes are needed. For example, supposing that the DNA/RNA hybrid sequence {"ACC","CCU"} is the fourth sequence in a sequence collection, then a bond index that refers to the "U" would be {4,2,3}.
- All DNA and RNA sequence letters can be connected with the "MultiHydrogen" bond type.
- In peptide sequences, not all bond types apply to all sequence chemicals. The following bond types can only connect the peptide letters shown:
-
"DisulfideBridges" C ↔ C, U ↔ U, C ↔ U "LactamBridges" D ↔ K, E ↔ K - For example, the type in BioSequence["Peptide","CGGGU",Bond[{1,5},type]] can be "DisulfideBridges" but not "LactamBridges".
- Bonds for a motif sequence can also be entered in dot-bracket notation. This form represents the bonds of a sequence as a single string where each letter of the sequence corresponds to that position in the string. Valid characters for the bond string are either a period ("."), which represents no bond or parenthesis ("(" and ")"), or angle brackets ("<" and ">"), which represent nested bonded pairs. For example, the string "<((..>))." would be appropriate for a sequence nine letters long and would be equivalent to {Bond[{1,6}],Bond[{2,8}],Bond[{3,7}]}.
- Properties "prop" of a BioSequence obtained by BioSequence[…]["prop"] include:
-
"SequenceType" the type of sequence as a "BioSequenceType" entity "SequenceString" a string representing the sequence "SequenceBondList" a list of all explicitly given bonds in the sequence "SequenceBondCount" number of explicitly given bonds in the sequence "SequenceLength" the length of the sequence "SequencePattern" a string expression expanding degenerate letters "AbbreviationSequence" a string representation using allowed abbreviations "ChemicalList" a list of the literal chemical entities "ChemicalPatternList" a list of entity patterns, allowing for degenerate letters "MolecularMass" the molecular mass of the sequence "MolarMass" the molar mass of the sequence "HELM" HELM string of the sequence "Properties" a list of the properties - Both "ChemicalList" and "ChemicalPatternList" give the particular chemicals for each term of the sequence. The former does not support degenerate letters, while the latter will represent them using Alternatives.
- If the sequence has degenerate terms, its molecular mass may be an Interval.
- The "HELM" property gives the Hierarchical Editing Language for Macromolecules (HELM) representation of the BioSequence.
- The types available to BioSequence can also be extended by creating an EntityStore with "ExtendedBioSequenceType" entities and then registering it (EntityRegister).
- The following "ExtendedBioSequenceType" properties can be defined:
-
"Alphabet" a list of the letters permitted within this sequence "AlphabetRules" an association from letters to specific chemicals "BibliographicSource" an external identifier documenting the sequence type "Caption" the caption above the sequence in formatted output "ComplementLetterRules" two-way rules defining a complement operation "Icon" the icon displayed in the formatted output of the sequence "MolecularMassRules" an association from letters to molecular masses - The "Icon" can be provided as either an image or the canonical name of an existing sequence type.
- The "MolecularMassRules" will override the molecular masses of the chemicals given via "AlphabetRules" and allow masses to be calculated when no chemicals are given.
- BioSequenceQ[bioseq] gives True only if bioseq corresponds to a valid BioSequence expression.
Examples
open all close allScope (28)
Basic Sequences (8)
Represent a circular DNA sequence:
Represent a circular RNA sequence:
Represent a circular peptide sequence:
Infer the type from the sequence of letters:
Specify a peptide sequence using standard abbreviations:
Infer the type of the sequence from standard abbreviations:
Degenerate terms can be entered as alternatives in a string expression:
Sequences from Entities (4)
Represent a sequence through a list of corresponding chemicals:
Degenerate letters can be specified by alternatives between chemicals:
Represent the DNA sequence of the BRCA1 gene:
Represent the peptide sequence of the protein myoglobin:
"BioSequenceType" entities can be used as the type when constructing biomolecular sequences:
Sequences with Bonds (4)
Bond can be used to add additional structure to the sequence:
The bond type does not need to be specified and will be inferred when needed and if possible:
Bonds in RNA can be specified using basic dot-bracket notation:
Hybrid Strands (5)
Hybrid strands are strands with multiple types of sequences bonded along their primary structure:
Motif-type inference works within hybrid strands:
Bonds can cross the motif sequences of a hybrid strand:
Bonds at the hybrid level can refer to a connection in a given motif:
Bonds can also be specified on the motif sequences of hybrid strands:
Sequence Collections (7)
Sequence collections represent a set of disconnected sequences unless additional bonds are provided:
Motif sequences can be connected by bonds at the sequence level:
Sequence collections can contain any mixture of motif and hybrid strands:
Type inference works on both the hybrid and motif strands in sequence collections:
Bonds can connect multiple hybrid strands:
Bonds can be specified on multiple levels in a sequence collection:
Represent a sequence collection with peptide and circular peptide components:
Generalizations & Extensions (1)
Properties & Relations (28)
BioSequence provides a number of properties:
The types of BioSequence are entities that contain many further properties describing the sequence:
Access the raw sequence string:
Find the length of the underlying sequence:
Resolve degenerate letters into patterns over specific bases:
Obtain a raw sequence string composed of abbreviations:
Specific sequences can be resolved into lists of chemicals:
Degenerate letters can be resolved into chemical alternatives:
Access the oligonucleotide (i.e. single-strand) molecular mass varying by possible degenerate choices:
The range of molar mass is also available for sequences with degenerate letters:
Obtain the HELM representation of a sequence:
Define a sequence type with molecular mass rules and a custom icon:
With defined mass rules, the molecular mass can be calculated:
Most properties of hybrid strands are lists of the properties of the underlying motif sequences:
Most properties of sequence collections are lists of lists of the underlying motif sequences:
The "MolecularMass" and "MolarMass" properties apply to hybrid strands as a whole:
Mass properties also apply to sequence collections as a whole:
The basic letters for a given type correspond to the "Alphabet" property of "BioSequenceType" entities:
BioSequence motifs can be provided as input to Molecule:
A hybrid strand BioSequence can also be given as an input to Molecule:
A BioSequence collection can also be provided to Molecule:
Use ConnectedMoleculeComponents to obtain the separate molecules of a sequence collection:
SequenceAlignment can find alignments between two instances of BioSequence:
RandomInstance can sample fully specified instances from a degenerate BioSequence:
BioSequenceQ can validate that a BioSequence is of a given type or has other attributes:
BioSequenceComplement and BioSequenceReverseComplement find genetic complements of a BioSequence:
BioSequencePlot shows a schematic diagram of a BioSequence:
When converting a BioSequence of type "DNA", "RNA", "CircularDNA" or "CircularRNA" to a Molecule, the sequence is interpreted to be going from the 5' 3' direction (positive-sense):
When converting a BioSequence of type "Peptide" or "CircularPeptide" to a Molecule, the sequence is interpreted to be going from the N-terminus to the C-terminus:
Possible Issues (4)
Sequences containing letters not defined for the given type will not format:
Subsequent operations with these sequences may not evaluate:
It may not be possible to infer a type of sequence appropriate for the given string:
Not all hybrid strands can be converted to Molecule:
Incompatible motif types in hybrid strands will also lead to no interpretation for the mass properties:
Standard abbreviations are not defined for all DNA and RNA letters:
Neat Examples (3)
Compare two very similar genes:
Generate sequences containing all of the supported characters:
Represent human insulin as a BioSequence:
Convert to a Molecule:
See Also
BioSequenceQ BioSequenceComplement BioSequenceReverseComplement BioSequenceTranslate BioSequenceBackTranslateList BioSequenceTranscribe BioSequenceModify BioSequenceInstances BioSequencePlot
Entity Types: BioSequenceType
Function Repository: BioSequenceGraph BioSequenceToRegularExpression BioSequenceMoleculePlot BioSequenceFoldingMatrix GetPubMedIdentifiers PeptideName NucleotideName NucleotideStaveDiagram NuIDEncode NuIDDecode RemoveDegenerateSequenceDifferences RNAFoldingMaximumBasePairing
Text
Wolfram Research (2020), BioSequence, Wolfram Language function, https://reference.wolfram.com/language/ref/BioSequence.html (updated 2022).
CMS
Wolfram Language. 2020. "BioSequence." Wolfram Language & System Documentation Center. Wolfram Research. Last Modified 2022. https://reference.wolfram.com/language/ref/BioSequence.html.
APA
Wolfram Language. (2020). BioSequence. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/BioSequence.html
BibTeX
@misc{reference.wolfram_2025_biosequence, author="Wolfram Research", title="{BioSequence}", year="2022", howpublished="\url{https://reference.wolfram.com/language/ref/BioSequence.html}", note=[Accessed: 18-February-2026]}
BibLaTeX
@online{reference.wolfram_2025_biosequence, organization={Wolfram Research}, title={BioSequence}, year={2022}, url={https://reference.wolfram.com/language/ref/BioSequence.html}, note=[Accessed: 18-February-2026]}