GeneralizedLinearModelFit
GeneralizedLinearModelFit[{{x1,y1},{x2,y2},…},{f1,f2,…},x]
constructs a generalized linear model of the form ![]() that fits the yi for each xi.
that fits the yi for each xi.
GeneralizedLinearModelFit[data,{f1,f2,…},{x1,x2,…}]
constructs a generalized linear model of the form ![]() where the fi depend on the variables xk.
where the fi depend on the variables xk.
GeneralizedLinearModelFit[{m,v}]
constructs a generalized linear model from the design matrix m and response vector v.
Details and Options




- GeneralizedLinearModelFit attempts to model the input data using a linear combination of functions transformed by a generic invertible function (link function).
- GeneralizedLinearModelFit produces a generalized linear model of the form
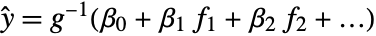 under the assumption that the original
under the assumption that the original 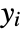 are independent observations following an exponential family distribution with mean
are independent observations following an exponential family distribution with mean 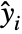 and the function
and the function 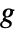 being an invertible link function.
being an invertible link function. - The ExponentialFamily option controls the distribution while the LinkFunction option controls the form of
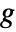 .
. - GeneralizedLinearModelFit returns a symbolic FittedModel object to represent the generalized linear model it constructs. The properties and diagnostics of the model can be obtained from model["property"].
- The value of the best-fit function from GeneralizedLinearModelFit at a particular point x1, … can be found from model[x1,…].
- Possible forms of data are:
-
{y1,y2,…} equivalent to the form {{1,y1},{2,y2},…} {{x11,x12,…,y1},…} a list of independent values xij and the responses yi {{x11,x12,…}y1,…} a list of rules between input values and responses {{x11,x12,…},…}{y1,y2,…} a rule between a list of input values and responses {{x11,…,y1,…},…}n fit the nth column of a matrix - With multivariate data such as
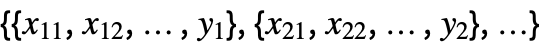 , the number of coordinates xi1, xi2, … should equal the number of variables xi.
, the number of coordinates xi1, xi2, … should equal the number of variables xi. - Additionally, data can be specified using a design matrix without specifying functions and variables:
-
{m,v} a design matrix m and response vector v - In GeneralizedLinearModelFit[m,v], the design matrix m is formed from the values of basis functions fi at data points in the form {{f1,f2,…},{f1,f2,…},…}. The response vector v is the list of responses {y1,y2,…}.
- For a design matrix m and response vector v, the model is
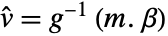 , where
, where 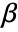 is the vector of parameters to be estimated.
is the vector of parameters to be estimated. - When a design matrix is used, the basis functions fi can be specified using the form GeneralizedLinearModelFit[{m,v},{f1,f2,…}].
- GeneralizedLinearModelFit takes the following options:
-
AccuracyGoal Automatic the accuracy sought ConfidenceLevel 95/100 confidence level for parameters and predictions CovarianceEstimatorFunction "ExpectedInformation" estimation method for the parameter covariance matrix DispersionEstimatorFunction Automatic function for estimating the dispersion parameter ExponentialFamily Automatic exponential family distribution for y IncludeConstantBasis True whether to include a constant basis function LinearOffsetFunction None known offset in the linear predictor LinkFunction Automatic link function for the model MaxIterations Automatic maximum number of iterations to use NominalVariables None variables considered as nominal PrecisionGoal Automatic the precision sought Weights Automatic weights for data elements WorkingPrecision Automatic the precision for internal computations - With the setting IncludeConstantBasis->False, a model of the form
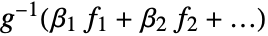 is fitted.
is fitted. - With the setting LinearOffsetFunction->h, a model of the form
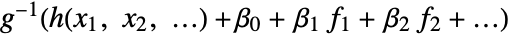 is fitted.
is fitted. - With ConfidenceLevel->p, probability-p confidence intervals are computed for parameter and prediction intervals.
- With the setting DispersionEstimatorFunction->f, the common dispersion is estimated by f[y,
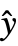 ,w] where y={y1,y2,…} is the list of observations,
,w] where y={y1,y2,…} is the list of observations, 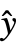 ={
={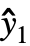 ,
,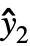 ,…} is the list of predicted values, and w={w1,w2,…} is the list of weights for the measurements yi.
,…} is the list of predicted values, and w={w1,w2,…} is the list of weights for the measurements yi. - Possible settings for ExponentialFamily include: "Gaussian", "Binomial", "Poisson", "Gamma", "InverseGaussian", or "QuasiLikelihood".
- Properties related to data and the fitted function obtained using model["property"] include:
-
"BasisFunctions" list of basis functions "BestFit" fitted function "BestFitParameters" parameter estimates "Data" the input data or design matrix and response vector "DesignMatrix" design matrix for the model "Function" best fit pure function "LinearPredictor" fitted linear combination "Response" response values in the input data "Weights" weights used to fit the data - Properties related to dispersion and model deviances include:
-
"Deviances" deviances "DevianceData" deviance table dataset "EstimatedDispersion" estimated dispersion parameter "NullDeviance" deviance for the null model "NullDegreesOfFreedom" degrees of freedom for the null model "ResidualDeviance" difference between the deviance for the fitted model and the deviance for the full model "ResidualDegreesOfFreedom" difference between the model degrees of freedom and null degrees of freedom - Types of residuals include:
-
"AnscombeResiduals" Anscombe residuals "DevianceResiduals" deviance residuals "FitResiduals" difference between actual and predicted responses "LikelihoodResiduals" likelihood residuals "PearsonResiduals" Pearson residuals "StandardizedDevianceResiduals" standardized deviance residuals "StandardizedPearsonResiduals" standardized Pearson residuals "WorkingResiduals" working residuals - Properties and diagnostics for parameter estimates include:
-
"CorrelationMatrix" asymptotic parameter correlation matrix "CovarianceMatrix" asymptotic parameter covariance matrix "ParameterEstimates" table of fitted parameter information - Properties related to influence measures include:
-
"CookDistances" list of Cook distances "HatDiagonal" diagonal elements of the hat matrix - Properties of predicted values include:
-
"PredictedResponse" fitted values for the data - Properties that measure goodness of fit include:
-
"AdjustedLikelihoodRatioIndex" Ben‐Akiva and Lerman's adjusted likelihood ratio index "AIC" Akaike Information Criterion "BIC" Bayesian Information Criterion "CoxSnellPseudoRSquared" Cox and Snell's pseudo 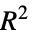
"CraggUhlerPseudoRSquared" Cragg and Uhler's pseudo 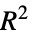
"EfronPseudoRSquared" Efron's pseudo 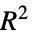
"LikelihoodRatioIndex" McFadden's likelihood ratio index "LikelihoodRatioStatistic" likelihood ratio "LogLikelihood" log likelihood for the fitted model "PearsonChiSquare" Pearson's 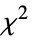 statistic
statistic
Properties
Examples
open allclose allBasic Examples (1)
Scope (15)
Data (8)
Fit data with success probability responses, assuming increasing integer-independent values:
Fit a model of more than one variable:
Fit data to a linear combination of functions of predictor variables:
Fit a rule of input values and responses:
Specify a column as the response:
Fit a model with categorical predictor variables:
Obtain a deviance table for the model:
Fit a model given a design matrix and response vector:
Fit the model referring to the basis functions as x and y:
Obtain a list of available properties for a generalized linear model:
Properties (7)
Data & Fitted Functions (1)
Residuals (1)
Dispersion and Deviances (1)
Parameter Estimation Diagnostics (1)
Influence Measures (1)
Prediction Values (1)
Generalizations & Extensions (1)
Options (10)
ConfidenceLevel (1)
The default gives 95% confidence intervals:
Set the level to 90% within FittedModel:
CovarianceEstimatorFunction (1)
DispersionEstimatorFunction (1)
ExponentialFamily (1)
IncludeConstantBasis (1)
LinearOffsetFunction (1)
Fit data to a canonical gamma regression model:
Fit data to a gamma regression model with a known Sqrt[x] term:
LinkFunction (1)
NominalVariables (1)
WorkingPrecision (1)
Use WorkingPrecision to get higher precision in parameter estimates:
Reduce the precision in property computations after the fitting:
Applications (2)
Properties & Relations (5)
DesignMatrix constructs the design matrix used by GeneralizedLinearModelFit:
By default, GeneralizedLinearModelFit and LinearModelFit fit equivalent models:
A default "Binomial" model is equivalent to the model for LogitModelFit:
ProbitModelFit is equivalent to a "Binomial" model with "ProbitLink":
GeneralizedLinearModelFit will use the time stamps of a TimeSeries as variables:
Rescale the time stamps and fit again:
GeneralizedLinearModelFit acts pathwise on a multipath TemporalData:
Text
Wolfram Research (2008), GeneralizedLinearModelFit, Wolfram Language function, https://reference.wolfram.com/language/ref/GeneralizedLinearModelFit.html.
CMS
Wolfram Language. 2008. "GeneralizedLinearModelFit." Wolfram Language & System Documentation Center. Wolfram Research. https://reference.wolfram.com/language/ref/GeneralizedLinearModelFit.html.
APA
Wolfram Language. (2008). GeneralizedLinearModelFit. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/GeneralizedLinearModelFit.html