SpatialJ
SpatialJ[pdata,r]
estimates the ![]() function
function ![]() for point data pdata at radius r.
for point data pdata at radius r.
SpatialJ[pproc,r]
computes ![]() for the point process pproc.
for the point process pproc.
SpatialJ[bdata,r]
computes ![]() for binned data bdata.
for binned data bdata.
SpatialJ[pspec]
generates the function ![]() that can be applied repeatedly to different radii r.
that can be applied repeatedly to different radii r.
Details and Options

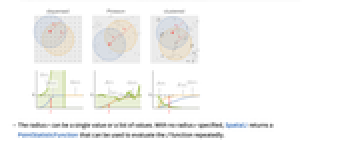


- The function
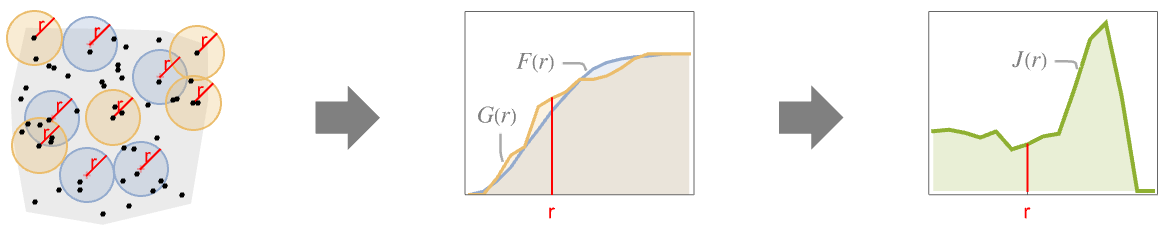
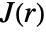 is the ratio of the probability of finding no point within distance
is the ratio of the probability of finding no point within distance 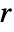 of a typical point to the probability of finding no point within distance
of a typical point to the probability of finding no point within distance 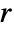 of any location. It is given as
of any location. It is given as 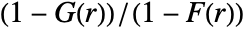 , where
, where 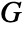 is NearestNeighborG and
is NearestNeighborG and 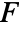 is EmptySpaceF.
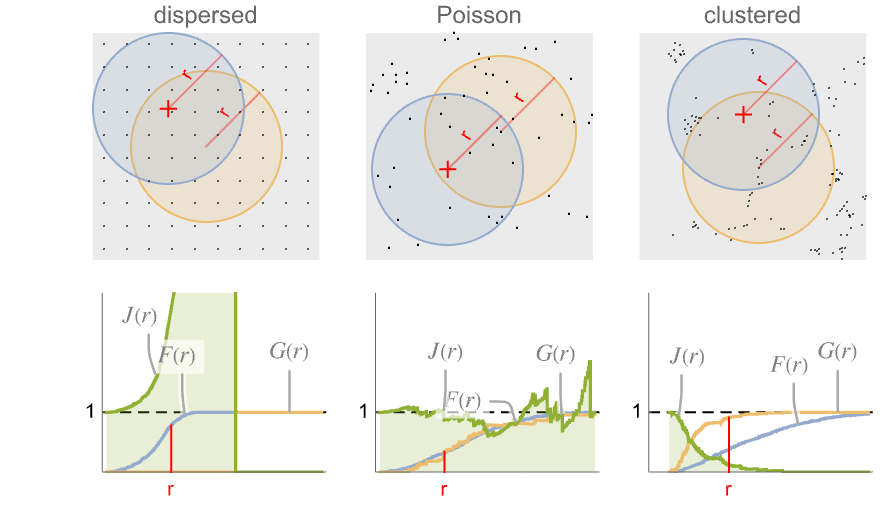
is EmptySpaceF. - SpatialJ measures spatial homogeneity of a point collection within distance
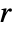 . In comparing with a Poisson point process:
. In comparing with a Poisson point process: -
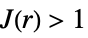
more dispersed than Poisson 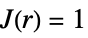
like Poisson, i.e. complete spatial randomness 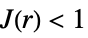
more clustered than Poisson - The radius r can be a single value or a list of values. With no radius r specified, SpatialJ returns a PointStatisticFunction that can be used to evaluate the
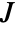 function repeatedly.
function repeatedly. - The points pdata can have the following forms:
-
{p1,p2,…} points pi GeoPosition[…],GeoPositionXYZ[…],… geographic points SpatialPointData[…] spatial point collection {pts,reg} point collection pts and observation region reg - If the observation region reg is not given, a region is automatically computed using RipleyRassonRegion.
- The point process pproc can have the following forms:
-
proc a point process proc {proc,reg} a point process proc and observation region reg - The observation region reg should be parameter free and SpatialObservationRegionQ.
- The binned data bdata is from SpatialBinnedPointData and is treated as an InhomogeneousPoissonPointProcess with a piecewise constant intensify function.
- For pdata,
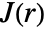 is computed from the points pi by combining the estimates of
is computed from the points pi by combining the estimates of 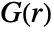 and
and 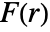 . The estimation assumes the point pattern is stationary in space.
. The estimation assumes the point pattern is stationary in space. - For pproc,
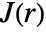 is computed by using exact formulas or by simulation to generate point data.
is computed by using exact formulas or by simulation to generate point data. - The following options can be given:
-
Method Automatic what methods to use SpatialBoundaryCorrection Automatic what boundary correction to use - The following settings can be used for SpatialBoundaryCorrection:
-
Automatic automatically determined boundary correction None no boundary correction "BorderMargin" use interior margin for observation region "Hanisch" drops points for which the distance to the nearest neighbor is greater than the distance to boundary "KaplanMeier" SurvivalDistribution method: the point distance to its nearest neighbor is censored by its distance to the region boundary "NelsonAalen" SurvivalDistribution method: the point distance to its nearest neighbor is censored by its distance to the region boundary - The setting Method->{"Discretization"->opts} allows for adjusting the discretization method in the estimation. Here opts can be any valid options for DiscretizeRegion.
Examples
open allclose allBasic Examples (3)
Estimate the ![]() function at a given distance:
function at a given distance:
Estimate the ![]() function within a range of distances:
function within a range of distances:
Visualize the result with ListPlot:
Scope (7)
Point Data (4)
Estimate the ![]() function for some data at distance 0.1:
function for some data at distance 0.1:
Obtain empirical estimates of the ![]() function for a list of given distances:
function for a list of given distances:
Create a PointStatisticFunction for future use:
Compute a value at a given radius:
Estimate the ![]() function without explicitly providing the observation region:
function without explicitly providing the observation region:
Observation region generated by Ripley–Rasson estimator:
Estimated ![]() function at distance 0.05:
function at distance 0.05:
Use SpatialJ with GeoPosition:
Point Processes (3)
The ![]() function function for PoissonPointProcess is always identity:
function function for PoissonPointProcess is always identity:
This is so because EmptySpaceF and NearestNeighborG of a PoissonPointProcess are identical:
The ![]() function for a cluster process ThomasPointProcess with specified dimension:
function for a cluster process ThomasPointProcess with specified dimension:
The ![]() function for a cluster process MaternPointProcess with specified dimension:
function for a cluster process MaternPointProcess with specified dimension:
Options (2)
SpatialBoundaryCorrection (1)
The SpatialJ estimator without boundary correction is biased and should not be used unless with a large point set:
The default method "BorderMargin" only considers the points that are distance ![]() from the boundary:
from the boundary:
The "Hanisch" method weights each point in the observation region to make the estimated values unbiased:
The "KaplanMeier" and "NelsonAalen" methods are estimators used in SurvivalDistribution. The distance of each point to its nearest neighbor point is censored by the distance of each point to the boundary of the observation region:
Method (1)
Discretization setting can be provided under Method as suboptions:
Estimate the ![]() function at the same radius with different values of MaxCellMeasure:
function at the same radius with different values of MaxCellMeasure:
Use different discretization methods to estimate the ![]() function at the same radius:
function at the same radius:
Applications (2)
Properties & Relations (3)
SpatialJ measures clustering of a point collection. ![]() indicates the points are clustered within distance
indicates the points are clustered within distance ![]() , while
, while ![]() indicates the points are dispersed within distance
indicates the points are dispersed within distance ![]() . Generate samples from lattice points, a Poisson point process and a Thomas point process:
. Generate samples from lattice points, a Poisson point process and a Thomas point process:
Estimate ![]() from each sample and compare the results:
from each sample and compare the results:
Visualize how NearestNeighborG impacts SpatialJ:
NearestNeighborG estimates the probability of finding another point within distance r from a point in the point collection:
Visualize how EmptySpaceF impacts SpatialJ:
EmptySpaceF estimates the probability of finding another point within distance r from a reference point:
Text
Wolfram Research (2020), SpatialJ, Wolfram Language function, https://reference.wolfram.com/language/ref/SpatialJ.html.
CMS
Wolfram Language. 2020. "SpatialJ." Wolfram Language & System Documentation Center. Wolfram Research. https://reference.wolfram.com/language/ref/SpatialJ.html.
APA
Wolfram Language. (2020). SpatialJ. Wolfram Language & System Documentation Center. Retrieved from https://reference.wolfram.com/language/ref/SpatialJ.html